Cite as: Leliaert F, Verbruggen H, Vanormelingen P, Steen F, Lopez-Bautista JM, Zuccarello GC & De Clerck O (2014) DNA-based species delimitation in algae. European Journal of Phycology 49: 179-196. pdf
Abstract. Given problems of species delimitation in algae using morphology or sexual compatibility, molecular data are becoming the standard for delimiting algal species and testing traditional species boundaries. The idea that species are separately evolving metapopulation lineages, along with theoretical progress in phylogenetic and population genetic analyses, has led to the development of new methods of species delimitation. We review these recent developments in DNA-based species delimitation methods, and discuss how they have and will continue to change our understanding of algal species boundaries. Although single-locus approaches have proven effective for a first rapid and large-scale assessment of species diversity, species delimitation based on single gene trees falls short due to gene tree – species tree incongruence, caused by confounding processes like incomplete lineage sorting, trans-species polymorphism, hybridisation and introgression. Data from unlinked loci and multispecies coalescent methods, which combine principles from phylogenetics and population genetics, may now be able to account for these complicating factors. Several of these methods also provide statistical support regarding species boundaries, which is important because speciation is a process, and therefore uncertainty about the precise species boundaries is inevitable in recently diverged lineages. Although algae are the primary focus in this paper, the general principles are easily transferable to other eukaryotes.
Introduction
The idea that biological nature is aggregated in discrete entities named species is widespread but not unanimously accepted (Mayr, 1996; De Queiroz, 2007; Barraclough, 2010). Most biologists agree that the species represents a fundamental category of biological organisation. Yet, they have debated endlessly over an all-encompassing definition and the criteria used to delimit species (Hey, 2006; Wilkins, 2009) and phycologists are no exception to this (Guiry, 1992; John & Maggs, 1997; Mann 1999, 2010).
Disagreement over the nature of species and their defining properties has not dissuaded phycologists from documenting algal diversity and describing new species. A screening of papers published in European Journal of Phycology and Phycologia in 2012 and 2013 revealed that half (68 out of 137 research papers; Supplementary Table S1) describe new taxa or make explicit statements on species boundaries. In virtually all of these papers species delimitation is based on molecular data combined with at least some information about the morphology, ecology or (eco-)physiology of the organisms. It seems that at least in the peer-reviewed literature, it has become rare for extant species to be defined solely by morphology. In some algal groups (e.g. diatoms) the majority of new taxa, however, are still being described on morphology in book series, such as Bibliotheca and Iconographia Diatomologica (Kusber & Jahn, 2003).
Our survey of papers also revealed that whereas molecular and phylogenetic techniques are usually explained in detail, the criteria or methods used to delimit species based on sequence data are rarely specified. Typically, a phylogeny containing multiple specimens per species is constructed, and species are delimited based on topological criteria: monophyly and distinctness from sister species expressed as branch lengths, and some measure of support (bootstrap most commonly). Alternatively, but following a similar rationale, species are delimited based on a ratio of intra- versus interspecific divergence (the so-called barcoding gap) (Hebert et al., 2004). Sometimes, other thresholds, such as the presence of compensatory base pair changes (CBCs) in the secondary structure of the ribosomal RNA internal transcribed spacer (ITS), are used as a proxy for species divergence (Coleman, 2009). In several of the surveyed papers, intraspecific divergence could not be estimated because only a single representative per species was included, and comparable interspecific branch lengths or p-distances are taken as evidence for species boundaries. Another often used criterion is concordance between independent data sources, when individuals belong to the same clades in phylogenies based on different molecular markers, or when there is morphological divergence or sexual incompatibility between lineages in a phylogeny. In the two journals surveyed, we did not find systematic differences in how species delimitation was approached in either macroalgae or microalgae.
It is relevant to ask the question whether and how current practices of species delimitation reflect on our perceived knowledge of algal species diversity. For example, if two individuals share identical 18S rDNA sequences, does this make them conspecific? Is it possible for two individuals lacking CBCs in their ITS2 sequences to represent different biological species? By extension, is reproductive isolation a prerequisite for species delimitation and do species need to be monophyletic with all markers chosen? These and other questions highlight an important gap in our understanding of what algal species are and how to delimit them.
In this review we first aim to address species delimitation from a conceptual perspective, followed by an overview of the common practices in the phycological literature, and finally we look ahead to see how new developments in sequencing technologies, and advances in phylogenetic and population genetic approaches are taking centre stage in species delimitation. Although algae are the primary focus in this paper, the general principles are easily transferable to other eukaryotic taxa.
Species concepts and species delimitation
In the years following the publication of Darwin’s “On the Origin of Species”, the emerging evolutionary biological community struggled to reconcile the gradual process of evolution with the discrete species that it appeared to have produced (Coyne & Orr, 2004). The Modern Evolutionary Synthesis brought a better understanding of how species evolve, and the view that species are not just arbitrary divisions of an evolutionary continuum, but discrete entities gained broader acceptance (Shaw, 1998). Ernst Mayr, amongst others, regarded a species as a group of interconnected populations that form an extended gene pool (metapopulation), or incorporating a time dimension, an ancestral-descendant sequence of metapopulations (metapopulation lineage) evolving separately from others, and thus having its own evolutionary tendencies and fate (Coyne & Orr, 2004). The application of this so called “evolutionary species concept” (Simpson, 1951), however, was problematic because it was purely theoretical and at that time of little practical use for species delimitation. The search for operational criteria to delimit species has led to a proliferation of species concepts, each based on different biological properties of a species (Mayden, 1997; Hausdorf, 2011).
The morphological species concept uses discontinuities in morphological variation to distinguish species. This concept has, for practical reasons, dominated algal systematics for many decades (John & Maggs, 1997; Mann, 1999). Although discontinuities in morphological variation will in many instances correspond to species boundaries, convergent morphological evolution, morphological stasis, phenotypic plasticity and polymorphism are common phenomena that limit the correspondence between how a species would be defined based on the morphological concept versus the evolutionary or biological species concepts (Verbruggen, 2014). Many algal species are known to exhibit substantial intraspecific morphological variation, either as a result of genetically controlled polymorphism or environmentally induced plasticity (e.g., de Senerpont Domis et al., 2003; Lurling, 2003; Logares et al., 2007). Ignoring this intraspecific morphological variation may result in an overestimation of species diversity (Trainor, 1998; Macaya & Zuccarello, 2010). On the other hand, morphological differences between two species will often only emerge after enough time has passed since lineage divergence, and therefore recently diverged species are likely to remain undetected (Fig. 1). Molecular phylogenetic data have indeed revealed numerous cases of (closely related) species that are morphologically indistinguishable (e.g., Zuccarello & West, 2003; De Clerck et al., 2005; Fraser et al., 2009; Fucikova et al., 2011; Gutner-Hoch & Fine, 2011; Degerlund et al., 2012; Kucera & Saunders, 2012; Moniz et al., 2012; Soehner et al., 2012; Payo et al., 2013; Souffreau et al., 2013). In addition, ancient cryptic lineages have been detected in algal groups that are morphologically depauperate, exhibit stabilizing selection, or are subject to convergent evolution towards reduced morphologies, such as planktonic unicells (Saez et al., 2003; de Vargas et al., 2004; Šlapeta et al., 2006; Krienitz & Bock, 2012; Škaloud & Rindi, 2013) and seaweeds (Verbruggen et al., 2009; Sutherland et al., 2011).

To overcome difficulties associated with morphological data, the biological species concept has been applied to assess species boundaries in some algal groups, including diatoms (Mann, 2010), and some red, green and brown algae (e.g., Coleman, 1977; Zuccarello & West, 2003; Hiraoka et al., 2011; Maggs et al., 2011). The biological species concept is closely allied to the evolutionary species concept and focuses on reproductive isolation to distinguish species. Although this concept has been widely embraced, many taxonomists grew dissatisfied with it because it did not give a clear indication of how strong reproductive isolation between populations had to be for them to qualify as different species. Reproductive barriers between sexually reproducing algal species are not always complete (Brodie et al., 1993; Coyer et al., 2002; Kamiya, 2004; Casteleyn et al., 2009; Zardi et al., 2011), and little is known about hybrid formation in most algal groups. Moreover, laboratory crosses also only take into account intrinsic reproductive barriers and not extrinsic barriers, such as differences in timing of sexual reproduction or ecological preferences. Finally, the application of the biological species concept is unfeasible or impractical in algae that reproduce only asexually, or for which sexual reproduction is difficult to induce under laboratory conditions.
The phylogenetic species concept was a strong rival to the biological species concept, and regards species as unit products of natural selection and descent that can be identified based on reciprocal monophyly and/or diagnosability (Mayden, 1997). Although monophyly and diagnosability can be based on any character (phenotypic or genotypic), it was the emergence of molecular data that made this species concept popular in algal systematics (Manhart & McCourt, 1992; John & Maggs, 1997).
Because the literature on species concepts is vast and outside the scope of this paper, we refer to Guiry (1992), Manhart & McCourt (1992), John & Maggs (1997) and Mann (2010) for comprehensive overviews of species concepts in algae. What is important here is that although many of the species concepts are related, none are completely satisfactory, and most are at least partially incompatible in that they can lead to different conclusions regarding species boundaries (Mallet, 1995).
Debates on species concepts may be reaching some sort of consensus though. There is fairly general agreement that species are principal units in biology, and that there is a common evolutionary idea underlying them: speciation results from isolation of populations by interrupted gene flow, resulting in divergence due to selection and drift, and ultimately in separately evolving metapopulation lineages (Coyne & Orr, 2004). As such, species will show greater evolutionary independence from one another than populations within a single species do, and this independence will be subject to the amount of gene flow between lineages and the time of lineage separation (Hey & Pinho, 2012). Several authors have argued that many of the competing species concepts actually agree on the basic notion that species are separately evolving metapopulation lineages (Mayden, 1997; De Queiroz, 2007). In their view, traditional species concepts become secondary species properties, rather than necessary properties of species (Fig. 1). When two lineages separate, they will eventually acquire different properties (e.g., become sexually incompatible or morphologically distinct), which may serve as diagnostic evidence relevant for species delimitation. Because during the process of speciation these secondary properties do not necessarily arise at the same time or in a regular order, or may not arise at all, conflict will occur between species concepts in many cases, especially between recently formed species.
It has become clear that species delimitation using morphological or breeding data is often problematic for the reasons discussed above. Molecular data are therefore particularly valuable for delimiting species and testing traditional species boundaries. Below, we provide an overview of recent developments in DNA-based sequence delimitation and discuss how these have shaped the current view on species boundaries in algae.
Gene genealogies and species phylogenies
Gene trees spanning intraspecific and interspecific evolution are vital to understanding the process of speciation (Templeton, 2001). Because the stochastic pattern of allelic coalescence can be modelled, it provides valuable information regarding lineage divergence, and hence species delimitation (Ence & Carstens, 2011). In phylogenetics, gene trees are often implicitly equated to species trees, but it is important to realize that population genetics processes are acting within the branches of a species tree (Pamilo & Nei, 1988; Avise & Wollenberg, 1997; Maddison, 1997) (Fig. 2). These processes, known as the coalescent, can have three important effects.
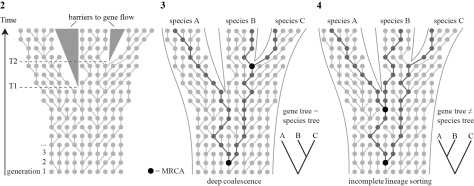
First, DNA divergence times may be considerably older than speciation times (deep coalescence, Fig. 3). Second, deep coalescence in combination with incomplete fixation of gene lineages within species lineages may result in gene trees and species trees having different topologies (incomplete lineage sorting, Fig. 4). Third, because incomplete lineage sorting is associated with the persistence of ancestral alleles in recently diverging lineages, gene trees will inevitably go through phases where individuals of a species are polyphyletic and paraphyletic before becoming monophyletic as alleles become fixed through time (Avise & Ball, 1990; Klein et al., 1998; Rosenberg & Nordborg, 2002) (Fig. 5). Clearly, this process has important consequences for delimitation of recently diverged species. Most importantly, it implies that reciprocal monophyly is not a necessary property of species, especially for recently diverged species (Kizirian & Donnelly, 2004; Knowles & Carstens, 2007). Simulation studies have indicated that a substantial amount of time (i.e. number of generations) may be required after lineage divergence before there will be a high probability of observing monophyly for a given locus, which is considered to depend on its effective population size (Avise, 2000; Hudson & Coyne, 2002; Hickerson et al., 2006).

Thus, although within-species gene genealogies differ in nature from among-species phylogenetic relationships (Posada & Crandall, 2001), young species reside in a boundary zone where both processes meet. Therefore, species delimitation should ideally incorporate aspects of both population genetics and phylogenetics (Knowles & Carstens, 2007; Hey & Pinho, 2012).
DNA-based species delimitation methods and application in algae
The idea that species are separately evolving metapopulation lineages, along with theoretical progress in phylogenetic and population genetic analyses has led to the development of new methods of species delimitation (Sites & Marshall, 2003; Camargo & Sites, 2013; Carstens et al., 2013). As discussed above, gene trees contain vital information regarding the speciation process, and hence DNA sequences are increasingly being used for species identification (DNA barcoding), species discovery and delimitation (DNA taxonomy), and for testing traditional species-level taxonomies (Wiens, 2007). Although the terms DNA taxonomy and DNA barcoding originally had different meanings (see glossary), the distinction between the two is not always clear-cut as both methodologies use similar molecular data (Collins & Cruickshank, 2013). For example, short, standardized gene regions (DNA barcodes) are increasingly being used for species discovery and delimitation. Similarly, DNA barcoding studies are not always restricted to analysis of short DNA sequences, but increasingly make use of multi-locus data (Roe et al., 2010).
For convenience, we subdivide DNA-based species delimitation methods into two categories, single-locus and multi-locus methods, although it should be noted that this subdivision is somewhat artificial as several methods can be applied to both single and multiple markers.
Single-locus species delimitation
Species delimitation methods based on single-locus data rely on the assumption that a single gene genealogy is representative of the species phylogeny. In general, single-locus methods impose a strict threshold of reciprocal monophyly for delimiting species and aim to detect discontinuities in sequence variation, assuming that interspecific divergence exceeds intraspecific variation.
Several single-locus methods for testing species boundaries are based on diagnostic character variation in DNA sequence data. These methods, which are rooted in the phylogenetic species concept, aggregate predefined populations with unique nucleotide differences into a single species (Davis & Nixon, 1992; Wiens & Penkrot, 2002). These methods have remained largely unexplored in algal studies because population boundaries in algae are often difficult to define a priori due to uncertainty regarding geographical ranges, and are therefore not further considered here.
Other methods do not require a priori definition of populations, and assess species boundaries based on molecular data alone. These methods rely on the assumptions that species are monophyletic for the gene under study and that there are discrete differences in sequence variation within and between species as a result of fixation of alleles in species lineages (Fig. 5). Distance approaches aim to detect a difference between inter- and intraspecific distances by analysing a frequency distribution of genetic distances between specimens. The transition between intra- and interspecific variation is visible as a distinct gap in the distribution, the so-called barcoding gap, and a corresponding distance threshold can then be applied to delimit species (Hebert et al., 2004). Similarly, tree-based methods seek to detect discontinuities in sequence variation that are associated with species boundaries by identifying clades of closely related sequences that are preceded by long and well supported branches. Distance and tree-based methods have been applied for identifying or delimiting species in several algal studies (e.g., Verbruggen et al., 2007; Rybalka et al., 2009; Freshwater et al., 2010).
Although in some cases clear discontinuities between intra- and interspecific genetic distances have been observed (e.g., Saunders, 2005; Zimmermann et al., 2011), the use of distance thresholds for species delineation may be problematic in some cases. First, among closely related species, levels of overlap between intra- and interspecific genetic distances are likely to be significant (Hamsher et al., 2011; Hoef-Emden, 2012). Second, with increased geographic sampling, barcoding gaps usually blur (Meyer & Paulay, 2005; Bittner et al., 2010; Bergsten et al., 2012). As a result, distance and tree-based methods often rely on intuition to decide whether sequence divergence is high enough or branches are long enough to consider a clade a distinct species.
Automated methods offering the promise of making single-locus species delimitation more efficient and less subjective have been developed. Software packages such as “Automatic Barcode Gap Discovery” (Puillandre et al., 2011) and “SPecies IDentity and Evolution” (Brown et al., 2012) aim at automatically detecting barcoding gaps in large datasets (Table 1). Another approach, based on statistical parsimony, separates groups of sequences into different networks if the haplotypes are connected by long branches that are affected by homoplasy (Hart & Sunday, 2007). This method has been applied in some algal groups, including the dinoflagellate Symbiodinium (Correa & Baker, 2009) and the seaweeds Boodlea and Padina (Leliaert et al., 2009; Silberfeld et al., 2013).
Table 1. Some algorithmic species delimitation methods and software packages.
| Method | Data | Brief description | Reference |
| ABGD | Single-locus | “Automatic Barcode Gap Discovery”. Distance methods that automates detection of a barcoding gap. | Puillandre et al., 2011 |
| Spider | Single-locus | “SPecies IDentity and Evolution”. Distance method assessing specimen identification efficacy based on a set of a priori defined species. Implemented in R, requires “ape”. | Brown et al., 2012 |
| Statistical parsimony | Single-locus | Partitions sequence data into independent networks of haplotypes connected by changes that are non-homoplastic with a 95% probability. | Templeton et al., 1992; Hart & Sunday, 2007 |
| GMYC model approach | Single-locus | Tree-based likelihood method fitting within- and between-species branching models to an ultrametric gene tree. Implemented in the R package SPLITS, requires ape (ape-package.ird.fr). | Pons et al., 2006; Monaghan et al., 2009 |
| bGMYC | Single-locus | Bayesian implementation of the GMYC model aimed at quantifying uncertainty in species limits by accounting for uncertainty in the phylogenetic tree and in the parameters of the model. Implemented in R, requires ape. | Reid & Carstens, 2012 |
| “Species delimitation” | Single-locus | Takes a priori defined species and a user-specific gene tree to compute the probability of the observed monophyly or exclusivity having occurred by chance in a coalescent process. Implemented as a plugin to Geneious (geneious.com). | Masters et al., 2011 |
| BPP | Multi-locus | Bayesian method estimating posterior distribution for species delimitation models. Uses a fixed species tree and prior information on population sizes and divergence times. | Yang & Rannala, 2010 |
| SpeDeSTEM | Multi-locus | Maximum likelihood method estimating the species tree for different models of species boundaries, which are compared with Akaike information criteria | Carstens & Dewey, 2010; Ence & Carstens, 2011 |
| O’Meara method | Multi-locus | Heuristic method that takes individual gene trees from multiple loci as input, and assesses differences between inter- and intraspecific branch length, and congruence among loci. Implemented in the Brownie package. | O’Meara, 2010 |
Likelihood methods based on evolutionary models have been proposed to statistically determine species boundaries based on analysis of branch lengths in phylogenetic trees. The idea behind these methods is that differences between species-level evolutionary processes and population-level evolutionary processes result in different branching rates in a gene tree. In a method proposed by Pons et al. (2006), a model that combines a coalescent model of intraspecific branching with a Yule model for interspecific branching (general mixed Yule-coalescent or GMYC model) is fitted to a gene tree, resulting in an estimation of species boundaries and a statistical measure of confidence for the species boundaries. The GMYC model approach has been refined to allow for a variable transition from coalescence to speciation among lineages (Monaghan et al., 2009), and to account for uncertainty in phylogenetic relationships and parameters of the GMYC model (Powell, 2012; Reid & Carstens, 2012; Fujisawa & Barraclough, 2014) (Table 1). The GMYC approach has been applied in a number studies on macro- and microalgae (Leliaert et al., 2009; Hoef-Emden, 2012; Tronholm et al., 2012; Payo et al., 2013; Silberfeld et al., 2013).
Marker choice for single-locus species delimitation
Irrespective of the species delimitation algorithm used, the success of single-locus species delimitation methods will depend on the history of species group under study (e.g., species delimitation will be more challenging in young lineages with some levels of inter-species gene flow than in older, well diverged lineages), the sampling strategy, and the molecular marker used. DNA sequences should ideally be obtained for a sufficient number of specimens and geographic locations to obtain a good estimate of intraspecific variation (Bergsten et al., 2012).
Apart from practical issues, such as ease of amplification, the effectiveness of a chosen neutral marker for molecular species discovery will depend on two criteria. First, the marker should contain sufficient variation within and among species. Second, interspecific variation should preferably exceed intraspecific variation to such a degree that a discontinuity in sequence variation, corresponding to the species boundary, is observable. This will depend on the coalescence time of the marker, which is determined by the effective population size and is independent of the mutation rate of the marker (Zink & Barrowclough, 2008).
Even though the mutation rate of a chosen marker will have no direct influence on its effectiveness to detect species, genetic variability is an important issue in DNA taxonomy. In principle, a single base pair difference could be sufficient for delimiting of a species (e.g., Brodie et al., 1996). However, species delimitation should ideally be based on more data because of the “strong claims require strong evidence” principle in science. Describing or delimiting a new species is a different thing and a stronger claim than identifying a specimen. Hence, more evidence is expected when delimiting species than for identifying specimens. Thus, species delimitation requires markers with fast coalescence (which depends on effective population size) and with sufficient variability (which depends on evolutionary rate and/or the length of the marker).
Given the prevalence of undiscovered and cryptic species in many algal groups, any marker chosen as a DNA barcode should preferably be suitable not only for species identification but also for species delimitation and discovery. For animals, the mitochondrial cytochrome oxidase I locus (COI or cox1) has been found to satisfy these criteria for most groups (Hebert et al., 2003) (but see Meier et al., 2006). However, COI has been shown to be less effective in many other eukaryotic groups (Pawlowski et al., 2012).
Because algae form a phylogenetically heterogeneous group, the application of a single universal marker for species delimitation is unfeasible, and different markers are applied for species delimitation in different algal groups. Table 2 summarizes the main markers that are currently used for species delimitation and/or DNA barcoding in different algal groups. It should be noted, however, that the resolution of these markers may not be optimal to discriminate between closely related species in specific clades (e.g., Mattio & Payri, 2010; Zardi et al., 2011; Janouškovec et al., 2013).
Species diversity surveys of microbial eukaryotic communities have heavily relied on the nuclear small subunit ribosomal RNA gene (SSU or 18S rDNA) (de Vargas et al., 1999; Moon-van der Staay et al., 2001; Not et al., 2012). Yet, comparison between genetic divergences of protein-coding vs. ribosomal RNA genes has indicated that the SSU rDNA generally underestimates the true number of species, especially in planktonic algae that have large population sizes and high turnover rates (Hall et al., 2010; Piganeau et al., 2011). Alternative universal markers have been proposed in various algal groups, including the nuclear and plastid rDNA (Sherwood & Presting, 2007; Liu et al., 2009).
Substitution rates of the Internal Transcribed Spacer (ITS) of the nuclear rDNA are much higher than those of the rRNA genes, and this region has been advocated as a marker for species level phylogenetics for macro- and microalgae (e.g., van der Strate et al., 2002; Lundholm et al., 2006; Gile et al., 2010; Pröschold et al., 2011). Unfortunately, several problems have been identified with the use of ITS for assessing species limits, including slow coalescence and intragenomic variation, potentially blurring the transition between species-level and population-level genetic distance (Álvarez & Wendel, 2003; Lane et al., 2007; Vanormelingen et al., 2008; Leliaert et al., 2009).
Special emphasis has been given to a particular kind of genetic distance threshold, which is found in the secondary structure of the ITS RNA product. Several studies have suggested that compensatory base changes (CBCs) in specific helices of the ITS correlate with reproductive isolation and could thus be used for delimiting species (Müller et al., 2007; Coleman, 2009; Wolf et al., 2013). Investigations of the correlation between species boundaries and CBCs suggest that although the presence of CBCs does correlate well with reproductive isolation barriers, the inverse is not necessarily true: absence of CBCs should therefore not be interpreted as proof for individuals belonging to a single species (Müller et al., 2007; Alverson, 2008; Caisová et al., 2011, 2013).
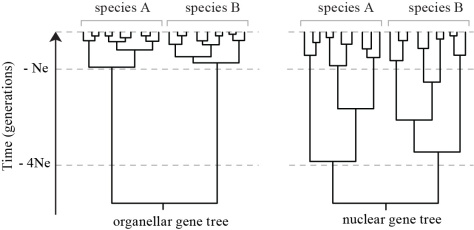
Species delimitation studies in many algal groups (macroalgae in particular) have largely relied on plastid and mitochondrial markers (Table 2). Besides a number of practical aspects that makes the amplification and sequencing of organellar loci relatively easy, their popularity for species delineation can be attributed to faster coalescence within species lineages compared to nuclear loci, resulting in clearer discontinuities between interspecific divergence and intraspecific variation (Palumbi et al., 2001) (Fig. 6). This expectation is congruent with population genetic theory, which predicts that for diploid organisms, the effective population size of nuclear DNA is four times higher than that of the haploid and uniparentally inherited organellar DNA (Hudson & Coyne, 2002). Although mitochondrial or chloroplast genes have been shown to segregate earlier during speciation than most nuclear genes and hence detect earlier stages of speciation (Lane et al., 2007; Birky, 2013; Payo et al., 2013), some authors have highlighted potential problems with organellar DNA and suggested that inferences about species limits, especially in recently diverging lineages, are unwarranted unless corroborated by nuclear gene data (Zink & Barrowclough, 2008).
Table 2. Main markers currently employed for DNA-based species delimitation and/or barcoding in some of the principal algal groups. Recommended barcode markers are indicated in bold, although it should be noted that for none of the groups a consensus has been reached.
| Algal group |
Marker |
References | ||
| plastid | mitochondrial | nuclear | ||
| Green algae | tufA, rbcL | SSU rDNA, LSU rDNA, rDNA ITS | Verbruggen et al., 2007; Leliaert et al., 2009; Hall et al., 2010; Luo et al., 2010; Saunders & Kucera, 2010; Škaloud & Peksa, 2010; Fucikova et al., 2011; Rindi et al., 2011; Škaloud & Rindi, 2013; Subirana et al., 2013 | |
| Red algae | rbcL, Rubiscospacer | cox1, cox2-3 spacer | Phycoerythrin, elongation factor, LSU rDNA | Robba et al., 2006; Sherwood et al., 2008; Le Gall & Saunders, 2010; Sherwood et al., 2010; Kucera & Saunders, 2012; Saunders & McDevit, 2012; Janouškovec et al., 2013; Payo et al., 2013 |
| Brown algae | psbA, rbcL, Rubisco spacer | cox1, cox3 | rDNA ITS | Lane et al., 2007; Kucera & Saunders, 2008; McDevit & Saunders, 2009; Mattio & Payri, 2010; Peters et al., 2010; Tronholm et al., 2010; Silberfeld et al., 2013 |
| Chrysophytes, Synurophytes | psaA, rbcL | cox1 | SSU rDNA, rDNA ITS | Boo et al., 2010; Kynčlová et al., 2010; Škaloud et al., 2012 |
| Cryptophytes | Rubisco spacer | cox1 | SSU rDNA, LSU rDNA, rDNA ITS | Lange et al., 2002; Hoef-Emden, 2007; Hoef-Emden, 2012 |
| Diatoms | rbcL | cox1 | SSU rDNA, LSU rDNA, rDNA ITS | Amato et al., 2007; Evans et al., 2007; Vanelslander et al., 2009; Mann et al., 2010; Moniz & Kaczmarska, 2010; Hamsher et al., 2011; Kermarrec et al., 2013 |
| Dinoflagellates | psbAncr, 23S rDNA | cox1, cob | LSU rDNA, rDNA ITS | Litaker et al., 2007; Lin et al., 2009; Litaker et al., 2009; Sampayo et al., 2009; Stern et al., 2010; LaJeunesse & Thornhill, 2011; Sato et al., 2011; LaJeunesse et al., 2012; Stern et al., 2012 |
| Haptophytes | tufA | cox1b-atp4 | SSU rDNA, LSU rDNA, rDNA ITS | Saez et al., 2003; de Vargas et al., 2004; Bendif et al., 2011; Edvardsen et al., 2011; Hagino et al., 2011; Sym et al., 2011; Bittner et al., 2013 |
| Raphidophytes | psaA, rbcL | cox1 | SSU rDNA, LSU rDNA, rDNA ITS | Demura et al., 2009; Klopper et al., 2013 |
| Xanthophytes | rbcL, psbA-rbcL spacer | rDNA ITS | Zuccarello & Lokhorst, 2005; Rybalka et al., 2009; Rybalka et al., 2013 | |
| Chlorarachniophytes | nuclear rDNA ITS, nucleomorph rDNA ITS | Gile et al., 2010 | ||
| Euglenophytes | SSU rDNA, LSU rDNA | SSU rDNA, LSU rDNA | Kim et al., 2013 | |
Multi-locus species delimitation
Although single-locus methods have proven effective for rapid and large-scale assessment of species diversity, concerns about the accuracy of species boundaries inferred from a single marker have been raised. Retention of ancestral polymorphism and incomplete lineage sorting, especially at early stages of speciation, will result in different neutral loci having their own gene trees that do not necessarily mirror the speciation process, with important consequences for assessing species boundaries (Hickerson et al., 2006; Knowles & Carstens, 2007). Furthermore, reticulate evolutionary processes such as hybridisation and introgression will remain unnoticed when using single gene data (e.g., Zardi et al., 2011; Mols-Mortensen et al., 2012). Although these criticisms have been vented ever since DNA taxonomy emerged, methods for multi-marker species delimitation have only recently been developed, mainly as a response to technical advances in sequencing technology that are making the collection of multi-marker datasets easier (Roe et al., 2010; Harrington & Near, 2012; Niemiller et al., 2012).
One approach uses genealogical concordance of unlinked, neutral loci to assess species boundaries. The rationale is that within species, the mixing effects of recombination between genes would cause unlinked loci to have different genealogies, but between isolated species, the extinction of ancestral alleles by genetic drift would lead to concordant genealogical histories. Hence, the transition between deep genealogical concordance (divergent genealogy) and shallow genealogical discordance (reticulate genealogy) can be used to delimit species (Avise & Ball, 1990; Baum & Shaw, 1995). In practice, species can thus be identified if gene genealogies of multiple unlinked loci show congruent patterns of reciprocal monophyly (Taylor et al., 2000; Dettman et al., 2003). The criteria of reciprocal monophyly and genealogical concordance of unlinked loci has been widely used (although not always explicitly) to delimit algal species (Casteleyn et al., 2008; Sherwood et al., 2008; Fraser et al., 2010; Škaloud & Peksa, 2010; Lundholm et al., 2012; Schmidt et al., 2012; Rybalka et al., 2013; Škaloud & Rindi, 2013), even if some of these studies showed possible signals of hybridization and/or introgression, blurring species boundaries (Zuccarello et al., 2005; Lane et al., 2007; Niwa et al., 2009; Destombe et al., 2010; Hind & Saunders, 2013).
Genealogical concordance of unlinked, neutral loci is expected to be present among well-diverged lineages. It should be noted that the majority of multi-locus species delimitation studies in algae show concordance between the different gene trees, and thus represent such well-diverged lineages. This is also suggested by molecular clock analyses, showing lineage divergences in the order of 100,000s to 10s of millions of years (Casteleyn et al., 2010; Payo et al., 2013; Souffreau et al., 2013), which appeared to have been sufficient to reach reciprocal monophyly across markers used in these studies. Between younger lineages however, as discussed above, the criteria of reciprocal monophyly and strict congruence will probably fail to detect species boundaries. New methods that extend coalescent models to the interspecific level (“multispecies coalescent”) offer perspectives for testing species boundaries in such shallow divergences (Table 1). The rationale of these methods is that despite species not necessarily being monophyletic in gene trees, a signal of species divergence may persist in gene trees of unlinked, neutral loci (Knowles & Carstens, 2007; O’Meara, 2010; Yang & Rannala, 2010).
Coalescence-based, multi-locus species delimitation methods are rapidly gaining popularity in studies on closely related species that are difficult to distinguish based on phenotypic features, and have been applied in various eukaryotic groups including animals (Carstens & Dewey, 2010; Leaché & Fujita, 2010; Yang & Rannala, 2010; Camargo et al., 2012), fungi (Leavitt et al., 2011) and land plants (Barrett & Freudenstein, 2011). Also in algae, phylogenetic and population genetic approaches have been combined to study boundaries between closely related species and assess confounding factors such as incomplete lineage sorting, trans-species polymorphism, hybridisation and introgression.
For example, Payo et al. (2013) used three unlinked loci from the mitochondrial, plastid and nuclear genome, and a multispecies coalescent method (BPP, Yang & Rannala, 2010) to infer 21 cryptic species of the red seaweed Portieria in the Philippines. The number of species identified using the BPP analysis was considerably higher than under the strict criteria of reciprocal monophyly and genealogical concordance across loci, with differences in species boundaries mainly situated in a clade uniting taxa that diverged at small spatial scales, probably during the Plio-Pleistocene. Such shallow divergences were captured only by the mitochondrial locus, which featured more rapid coalescence within species lineages compared to the plastid and nuclear loci.
Species boundaries have also been under close scrutiny in Fucus, a brown algal genus in which the taxonomy has been confounded by significant intraspecific morphological variability. Species boundaries in one of the main clades have remained largely unresolved, even in analyses of variable markers such as the nuclear rDNA ITS and a mitochondrial intergenic spacer, which was attributed to hybridization and/or incomplete lineage sorting, in what was believed to be a recent and rapid radiation within the last 3.8 million years (Serrao et al., 1999; Coyer et al., 2006). A multi-locus phylogenetic analysis of 13 nuclear loci (Canovas et al., 2011) provided finer resolution, resolving nearly all currently recognized species, including F. guiryi, confirming its recent elevation to species level based on an integrative approach employing phylogenetic and population genetic analyses (based on 14 SNPs and two microsatellite loci), along with morphological, physiological and distributional data (Zardi et al., 2011).
One of the diatom genera in which species boundaries have been thoroughly examined is Pseudo-nitzschia. In this genus, DNA sequence data are consistently used for species delimitation, which has resulted in the recent descriptions of numerous cryptic or pseudo-cryptic species (Amato et al., 2007; Lelong et al., 2012). In P. pungens, phylogenetic analysis has resulted in the recognition of three lineages that diverged recently, between 200 kya and 1.6 Mya (Casteleyn et al., 2010). These lineages differ by only a few bases in their ITS and rbcL sequences, but they were consistently recovered in both gene trees. Although the two most closely related clades were found to interbreed in laboratory crosses and form natural hybrids (Casteleyn et al., 2008; Casteleyn et al., 2009), sympatric populations from both clades were highly differentiated for microsatellites (Adams et al., 2009). Such differentiation independent of geography suggests the presence of some barrier to gene flow, and the presence of different species.
Future perspectives
Many taxonomists may feel uncomfortable with the idea of delimiting species based solely on DNA sequence data, and this is at least partially reflected in a reluctance to formally describe new algal species in many molecular studies (De Clerck et al., 2013). Several researchers have advocated that adequate delimitation of species should be based on additional lines of evidence, including morphological, ultrastructural, biochemical, geographical, ecological and/or breeding data (e.g., Pröschold et al., 2001; Vanormelingen et al., 2007; Walker et al., 2009; Bendif et al., 2011; McManus & Lewis, 2011; Neustupa et al., 2011; Ni-Ni-Win et al., 2011; Lam et al., 2012; Milstein & Saunders, 2012; Škaloud & Rindi, 2013). Although comparison of DNA-based species delimitation and species boundaries based on integrative taxonomy are useful in this respect, we argue that DNA sequence data serve as a reliable source of data for testing species boundaries even in the absence of additional phenotypic evidence. The hundreds of new species discovered using DNA sequences in the past 20 years have profoundly reshaped our ideas on algal diversity and present a telling case of our inability to accurately assess species diversity based on phenotypic characters alone. So far, the vast majority of those newly discovered lineages are not the result of recent speciation events that are difficult to detect due to a lack of variation in molecular markers and/or lineage sorting-related issues. Instead these species result from speciation events that predate the detection limit of most traditional molecular markers, often by millions of years.
The incessant flood of papers reporting new species based on DNA sequence data demonstrates that there are many more algal species remaining to be discovered and described. However, it would be naive to assume that the current (molecular) criteria to delimit species form an endpoint in the quest to describe the algal species diversity. On the one hand, our current procedures may result in species definitions that are too broad and encompass multiple separately evolving lineages. Conversely, they could lead to oversplitting if subclades within a species are erroneously considered to be separate species (Zachos & Lovari, 2013). These are both real issues, and further refinements in our understanding of algal diversity will require multi-marker datasets, and integration of population genetic and phylogenetic methods to disentangle complications caused by processes like incomplete lineage sorting, trans-species polymorphism, hybridisation and introgression (Fujita et al., 2012; Camargo & Sites, 2013).
Because speciation is a process and not a single event in time (Hey & Pinho, 2012), uncertainty about species boundaries is inevitable in recently diverged lineages. One of the strengths of DNA-based species delimitation is that this uncertainty can be taken into account and quantified. While the methods applying strict thresholds to delimit species (e.g. reciprocal monophyly, genetic distance, CBCs, or a barcoding gap) do not do this, several of the newly proposed methods incorporate probabilistic tests for species boundaries that provide statistical support plus a level of uncertainty regarding species boundaries (Fujita et al., 2012).
Carstens et al. (2013) have argued that although new DNA-based species delimitation methods hold great promise, it is important to be aware that they do make simplified assumptions about the process of speciation. Should these assumptions be violated in natural systems, suboptimal species limits may result. Because no model is ideal, Carstens et al. (2013) argue that species delimitation should preferably be based on a wide range of methods and a conservative consensus across methods. One important assumption made in coalescent-based species delimitation approaches is that data are sampled from neutral loci. However, it is well documented that divergent selection on ecological traits can lead to local adaptation and correlated reproductive isolation in a process of ecological speciation (Coyne & Orr, 2004; Camargo & Sites, 2013). In such cases, neutral markers would be uninformative for assessing rapid phenotypic divergence and speciation, necessitating the sampling of loci under selection that are potentially associated with selected traits (Nosil et al., 2009). Another assumption made in current coalescent-based species delimitation methods (BPP and spede-STEM) is that shared polymorphism is the result of incomplete lineage sorting. However, hybridisation and gene flow between species may be prevalent in algae, decreasing the accuracy of species delimitation (Camargo et al., 2012; Leaché et al., 2014).
While it is now generally recognized that multi-locus data are essential for accurate species delimitation (Dupuis et al., 2012), sampling of multiple unlinked genes using Sanger sequencing is challenging, especially because nuclear genes are essential. High-throughput sequencing methods are already facilitating the collection of multi-locus datasets for large numbers of individuals (McCormack et al., 2012). These technologies also offer perspectives for sampling new types of informative markers. For example a restriction-site-associated DNA (RAD-tag) sequencing approach has been developed to genotype genome-wide single-nucleotide polymorphisms (SNPs) (Baird et al., 2008), which are valuable for species delimitation studies in recent radiations and groups with occasional hybridization occurring between species (Shaffer & Thomson, 2007; Emerson et al., 2010; Wagner et al., 2013). Anchored hybrid enrichment or sequence capture is another development useful for producing large sets of nuclear loci across a wide range of samples (Lemmon et al., 2012), which will be highly informative about species boundaries (McCormack et al., 2013).
In conclusion, it is important to keep in mind the moving target that we are trying to circumscribe, a species. The question of species definition and delimitation is tied to the process of speciation, which has idiosyncratic components involving multiple populations, individuals with different life cycles and reproductive successes, different selective histories, and which contains a large component of stochasticity. Elucidating the relative importance of these components in the speciation process defines the beauty of systematic biology. The use of molecular markers in species delimitation has pointed phycologists toward more realistic species boundaries. We anticipate that multispecies coalescent methods based on multi-locus data will further refine our view on algal species, especially in recently diverged lineages, in which factors such as incomplete lineage sorting, hybridisation and introgression may confound species boundaries. These methods will increase the statistical rigour and objectivity of species delimitation, and are likely to result in the recognition of less inclusive entities, which in turn will have implications for estimates of algal species diversity. Resetting species boundaries towards a point where they truly reflect the biological reality will make the study of speciation processes more expedient. But quite likely, the question of “what is a species?” will remain with us as long as we want to study the process of evolution that produces these apparent discontinuities that we call “species”.
Acknowledgements. We thank David Mann for his invitation and encouragement to submit this review paper on DNA based species delimitation in algae. We are grateful to Juliet Brodie and Wilson Freshwater for their helpful remarks that helped us to improve the manuscript.
Glossary
- Ancestral polymorphism: genetic variation that originated prior to a speciation event. The presence of ancestral polymorphism in closely related species (shared ancestral polymorphism) may result in gene tree – species tree incongruence (Maddison & Knowles, 2006) (see also Trans-species polymorphism).
- Barcoding gap: a gap observable in the frequency distribution of intraspecific and interspecific genetic distances; a corresponding distance threshold may be used to delimit species.
- Biological species concept (BSC): a species concept that defines a species as “a group of interbreeding natural populations that are reproductively isolated from other such groups” (Mayr & Ashlock, 1991).
- Coalescence: looking back in time, the point at which two alleles converged on a single ancestral copy, known as the most recent common ancestor (MRCA) (Figs 2-5).
- Coalescent theory: “the mathematical and probabilistic theory underlying the evolutionary history of alleles” within a population lineage (Fujita et al., 2012).
- Deep coalescence: coalescence of alleles occurring significantly earlier than the divergence of the species containing those alleles (Fig. 3).
- DNA barcoding: a method of species identification, which identifies specimens based on DNA sequence similarity against a sequence database of a priori defined species (Hebert et al., 2003).
- DNA taxonomy: DNA-based species delimitation, which defines species boundaries based on sequence data (Tautz et al., 2003).
- Evolutionary species concept: a species concept that defines a species as “a lineage of ancestral descendant populations which maintains its identity from other such lineages and which has its own evolutionary tendencies and historical fate” (Wiley, 1978).
- General Mixed Yule Coalescent (GMYC) model: a model of a phylogenetic tree that separately considers branching within species (neutral coalescent model) and branching between species (Yule model).
- Incomplete lineage sorting (ILS): the maintenance of genetic variation within a metapopulation lineage from one speciation event to the next, resulting in deep coalescence and gene tree – species tree incongruence (Baum & Smith, 2012).
- Lineage sorting: the process by which alleles are inherited and lost over time.
- Metapopulation lineages: “Metapopulations” refers to a set of populations connected by gene flow over a relatively short time interval. “Lineage” refers to an ancestral-descendent sequence. Thus, a “metapopulation lineage” is a set of populations connected by gene flow on an evolutionary timescale (de Queiroz, 2005).
- Multispecies coalescent: coalescent model extended to the interspecific level, i.e. applied to gene trees in a species tree (Degnan & Rosenberg, 2009).
- Phylogenetic species concept: a species concept that defines species as “the smallest biological entities that are diagnosable and/or monophyletic” (Mayden, 1997).
- Reciprocal monophyly: monophyletic with respect to each other. Two sister taxa are reciprocally monophyletic when all alleles within each taxon are more closely related to one another than to any alleles in the other taxon.
- Trans-species polymorphism: presence of similar alleles in related species generated by the passage of alleles from ancestral to descendant species (Klein et al., 1998) (Fig. 5).
- Yule model: a birth-only model of a phylogenetic tree in which each branch is associated with a birth rate determining the rate at which the branch bifurcates into two branches.
References
Adams, N.G., Trainer, V.L., Rocap, G., Herwig, R.P., & Hauser, L. (2009). Genetic population structure of Pseudo-nitzschia pungens (Bacillariophyceae) from the Pacific Northwest and the North Sea. J. Phycol. 45: 1037-1045.
Álvarez, I. & Wendel, J.F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29: 417-434.
Alverson, A.J. (2008). Molecular systematics and the diatom species. Protist 159: 339-353.
Amato, A., Kooistra, W., Ghiron, J.H.L., Mann, D.G., Proschold, T., & Montresor, M. (2007). Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158: 193-207.
Avise, J. (2000). Phylogeography: the history and formation of species. Harvard University Press, Cambridge, Massachusetts.
Avise, J.C. & Ball, R.M. (1990). Principles of genealogical concordance in species concepts and biological taxonomy. Oxf. Surv. Evol. Biol. 7: 45-67.
Avise, J.C. & Wollenberg, K. (1997). Phylogenetics and the origin of species. Proc. Natl Acad. Sci. U.S.A. 94: 7748-7755.
Baird, N.A., Etter, P.D., Atwood, T.S., Currey, M.C., Shiver, A.L., Lewis, Z.A., Selker, E.U., Cresko, W.A., & Johnson, E.A. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376.
Barraclough, T.G. (2010). Evolving entities: towards a unified framework for understanding diversity at the species and higher levels. Phil. Trans. R. Soc. B 365: 1801-1813.
Barrett, C.F. & Freudenstein, J.V. (2011). An integrative approach to delimiting species in a rare but widespread mycoheterotrophic orchid. Mol. Ecol. 20: 2771-2786.
Baum, D.A. & Shaw, K.L. (1995). Genealogical perspectives on the species problem. In Experimental and Molecular Approaches to Plant Biosystematics (53) (Hoch, P.C. & Stephenson, A.G., editors), 289-303. Monographs in Systematic Botany from the Missouri Botanical Garden.
Baum, D.A. & Smith, S.D. (2012). Tree Thinking: An Introduction to Phylogenetic Biology. Roberts and Co., Greenwood Village.
Bendif, E.M., Probert, I., Herve, A., Billard, C., Goux, D., Lelong, C., Cadoret, J.-P., & Veron, B. (2011). Integrative taxonomy of the Pavlovophyceae (Haptophyta): A reassessment. Protist 162: 738-761.
Bergsten, J., Bilton, D.T., Fujisawa, T., Elliott, M., Monaghan, M.T., Balke, M., Hendrich, L., Geijer, J., Herrmann, J., Foster, G.N., Ribera, I., Nilsson, A.N., Barraclough, T.G., & Vogler, A.P. (2012). The effect of geographical scale of sampling on DNA barcoding. Syst. Biol. 61: 851-869.
Birky, C.W., Jr. (2013). Species detection and identification in sexual organisms using population genetic theory and DNA sequences. PLoS One 8: e52544.
Bittner, L., Gobet, A., Audic, S., Romac, S., Egge, E.S., Santini, S., Ogata, H., Probert, I., Edvardsen, B., & de Vargas, C. (2013). Diversity patterns of uncultured Haptophytes unravelled by pyrosequencing in Naples Bay. Mol. Ecol. 22: 87-101.
Bittner, L., Halary, S., Payri, C., Cruaud, C., de Reviers, B., Lopez, P., & Bapteste, E. (2010). Some considerations for analyzing biodiversity using integrative metagenomics and gene networks. Biol. Direct 5: 47.
Boo, S.M., Kim, H.S., Shin, W., Boo, G.H., Cho, S.M., Jo, B.Y., Kim, J.H., Yang, E.C., Siver, P.A., Wolfe, A.P., Bhattacharya, D., Andersen, R.A., & Yoon, H.S. (2010). Complex phylogeographic patterns in the freshwater alga Synura provide new insights into ubiquity vs. endemism in microbial eukaryotes. Mol. Ecol. 19: 4328-4338.
Brodie, J., Guiry, M.D., & Masuda, M. (1993). Life history, morphology and crossability of Chondrus ocellatus forma ocellatus and C. ocellatus forma crispoides (Gigartinales, Rhodophyta) from the north-western Pacific. Eur. J. Phycol. 28: 183-196.
Brodie, J., Hayes, P.K., Barker, G.L., & Irvine, L.M. (1996). Molecular and morphological characters distinguishing two Porphyra species (Rhodophyta: Bangiophycidae). Eur. J. Phycol. 31: 303-308.
Brown, S.D.J., Collins, R.A., Boyer, S., Lefort, M.-C., Malumbres-Olarte, J., Vink, C.J., & Cruickshank, R.H. (2012). Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 12: 562-565.
Caisová, L., Marin, B., & Melkonian, M. (2011). A close-up view on ITS2 evolution and speciation – a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol. Biol. 11: 262.
Caisová, L., Marin, B., & Melkonian, M. (2013). A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 164: 482-496.
Camargo, A., Morando, M., Avila, L.J., & Sites, J.W., Jr. (2012). Species delimitation with abc and other coalescent-based methods: A test of accuracy with simulations and an empirical example with lizards of the Liolaemus darwinii complex (Squamata: Liolaemidae). Evolution 66: 2834-2849.
Camargo, A. & Sites, J.J. (2013). Species delimitation: A decade after the renaissance. In The Species Problem – Ongoing Issues (Pavlinov, I., editor), 225-247. InTech, New York.
Canovas, F.G., Mota, C.F., Serrao, E.A., & Pearson, G.A. (2011). Driving south: a multi-gene phylogeny of the brown algal family Fucaceae reveals relationships and recent drivers of a marine radiation. BMC Evol. Biol. 11: 371.
Carstens, B.C. & Dewey, T.A. (2010). Species delimitation using a combined coalescent and information-theoretic approach: An example from North American Myotis bats. Syst. Biol. 59: 400-414.
Carstens, B.C., Pelletier, T.A., Reid, N.M., & Satler, J.D. (2013). How to fail at species delimitation. Mol. Ecol. 22: 4369-4383.
Casteleyn, G., Adams, N.G., Vanormelingen, P., Debeer, A.E., Sabbe, K., & Vyverman, W. (2009). Natural hybrids in the marine diatom Pseudo-nitzschia pungens (Bacillariophyceae): genetic and morphological evidence. Protist 160: 343-354.
Casteleyn, G., Chepurnov, V.A., Leliaert, F., Mann, D.G., Bates, S.S., Lundholm, N., Rhodes, L., Sabbe, K., & Vyverman, W. (2008). Pseudo-nitzschia pungens (Bacillariophyceae): A cosmopolitan diatom species? Harmful Algae 7: 241-257.
Casteleyn, G., Leliaert, F., Backeljau, T., Debeer, A.-E., Kotaki, Y., Rhodes, L., Lundholm, N., Sabbe, K., & Vyverman, W. (2010). Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc. Natl Acad. Sci. U.S.A. 107: 12952-12957.
Churro, C.I., Carreira, C.C., Rodrigues, F.J., Craveiro, S.C., Calado, A.J., Casteleyn, G., & Lundholm, N. (2009). Diversity and abundance of potentially toxic Pseudo-nitzschia Peragallo in Aveiro coastal lagoon, Portugal and description of a new variety, P. pungens var. aveirensis var. nov. Diatom. Res. 24: 35-62.
Coleman, A.W. (1977). Sexual and genetic isolation in cosmopolitan algal species Pandorina morum. Am. J. Bot. 64: 361-368.
Coleman, A.W. (2009). Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 50: 197-203.
Collins, R.A. & Cruickshank, R.H. (2013). The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 13: 969-975.
Correa, A. & Baker, A. (2009). Understanding diversity in coral-algal symbiosis: a cluster-based approach to interpreting fine-scale genetic variation in the genus Symbiodinium. Coral Reefs 28: 81-93.
Coyer, J.A., Hoarau, G., Oudot-Le Secq, M.P., Stam, W.T., & Olsen, J.L. (2006). A mtDNA-based phylogeny of the brown algal genus Fucus (Heterokontophyta; Phaeophyta). Mol. Phylogenet. Evol. 39: 209-222.
Coyer, J.A., Peters, A.F., Hoarau, G., Stam, W.T., & Olsen, J.L. (2002). Hybridization of the marine seaweeds, Fucus serratus and Fucus evanescens (Heterokontophyta : Phaeophyceae) in a 100-year-old zone of secondary contact. Proc. R. Soc. B 269: 1829-1834.
Coyne, J.A. & Orr, H.A. (2004). Speciation. Sinauer Associates, Sunderland, MA.
Darling, K.F. & Wade, C.A. (2008). The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Mar. Micropaleontol. 67: 216-238.
Davis, J.I. & Nixon, K.C. (1992). Populations, genetic-variation, and the delimitation of phylogenetic species. Syst. Biol. 41: 421-435.
De Clerck, O., Gavio, B., Fredericq, S., Barbara, I., & Coppejans, E. (2005). Systematics of Grateloupia filicina (Halymeniaceae, Rhodophyta), based on rbcL sequence analyses and morphological evidence, including the reinstatement of G. minima and the description of G. capensis sp. nov. J. Phycol. 41: 391-410.
De Clerck, O., Guiry, M.D., Leliaert, F., Samyn, Y., & Verbruggen, H. (2013). Algal taxonomy: A road to nowhere? J. Phycol. 49: 215-225.
de Queiroz, K. (2005). Ernst Mayr and the modern concept of species. Proc. Natl Acad. Sci. U.S.A. 102: 6600-6607.
De Queiroz, K. (2007). Species concepts and species delimitation. Syst. Biol. 56: 879-886.
de Senerpont Domis, L.N., Fama, P., Bartlett, A.J., Prud’homme van Reine, W.F., & Espinosa, C.A. (2003). Defining taxon boundaries in members of the morphologically and genetically plastic genus Caulerpa (Caulerpales, Chlorophyta). J. Phycol. 39: 1019-1037.
de Vargas, C., Norris, R., Zaninetti, L., Gibb, S.W., & Pawlowski, J. (1999). Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc. Natl Acad. Sci. U.S.A. 96: 2864-2868.
de Vargas, C., Sáez, A.G., Medlin, L.K., & Thierstein, H.R. (2004). Super-species in the calcareous plankton. In Coccolithophores: from molecular processes to global impact (Thierstein, H.R. & Young, J.R., editors), 271-298. Springer-Verlag, Berlin.
Degerlund, M., Huseby, S., Zingone, A., Sarno, D., & Landfald, B. (2012). Functional diversity in cryptic species of Chaetoceros socialis Lauder (Bacillariophyceae). J. Plankton Res. 34: 416-431.
Degnan, J.H. & Rosenberg, N.A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24: 332-340.
Demura, M., Noel, M.-H., Kasai, F., Watanabe, M.M., & Kawachi, M. (2009). Taxonomic revision of Chattonella antiqua, C. marina and C. ovata (Raphidophyceae) based on their morphological characteristics and genetic diversity. Phycologia 48: 518-535.
Destombe, C., Valero, M., & Guillemin, M.L. (2010). Delineation of two sibling red algal species, Gracilaria gracilis and Gracilaria dura (Gracilariales, Rhodophyta), using multiple DNA markers: Resurrection of the species G. dura previously described in the Northern Atlantic 200 years ago. J. Phycol. 46: 720-727.
Dettman, J.R., Jacobson, D.J., & Taylor, J.W. (2003). A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703-2720.
Dupuis, J.R., Roe, A.D., & Sperling, F.A.H. (2012). Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol. Ecol. 21: 4422-4436.
Edvardsen, B., Eikrem, W., Throndsen, J., Saez, A.G., Probert, I., & Medlin, L.K. (2011). Ribosomal DNA phylogenies and a morphological revision provide the basis for a revised taxonomy of the Prymnesiales (Haptophyta). Eur. J. Phycol. 46: 202-228.
Emerson, K.J., Merz, C.R., Catchen, J.M., Hohenlohe, P.A., Cresko, W.A., Bradshaw, W.E., & Holzapfel, C.M. (2010). Resolving postglacial phylogeography using high-throughput sequencing. Proc. Natl Acad. Sci. U.S.A. 107: 16196-16200.
Ence, D.D. & Carstens, B.C. (2011). SpedeSTEM: a rapid and accurate method for species delimitation. Mol. Ecol. Resour. 11: 473-480.
Evans, K.M., Wortley, A.H., & Mann, D.G. (2007). An assessment of potential diatom barcode genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158: 349-364.
Fraser, C.I., Hay, C.H., Spencer, H.G., & Waters, J.M. (2009). Genetic and morphological analyses of the southern bull kelp Durvillaea antarctica (Phaeophyceae: Durvillaeales) in New Zealand reveal cryptic species. J. Phycol. 45: 436-443.
Fraser, C.I., Winter, D.J., Spencer, H.G., & Waters, J.M. (2010). Multigene phylogeny of the southern bull-kelp genus Durvillaea (Phaeophyceae: Fucales). Mol. Phylogenet. Evol. 57: 1301-1311.
Freshwater, D.W., Tudor, K., O’Shaughnessy, K., & Wysor, B. (2010). DNA barcoding in the red algal order Gelidiales: comparison of COI with rbcL and verification of the “barcoding gap”. Cryptogam. Algol. 31: 435-449.
Fucikova, K., Rada, J.C., Lukesova, A., & Lewis, L.A. (2011). Cryptic diversity within the genus Pseudomuriella Hanagata (Chlorophyta, Chlorophyceae, Sphaeropleales) assessed using four barcode markers. Nova Hedwigia 93: 29-46.
Fujisawa, T. & Barraclough, T.G. (2014). Delimiting species using single-locus data and the generalized mixed yule coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Syst. Biol. 62: 707-724.
Fujita, M.K., Leaché, A.D., Burbrink, F.T., McGuire, J.A., & Moritz, C. (2012). Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 27: 480-488.
Gile, G.H., Stern, R.F., James, E.R., & Keeling, P.J. (2010). DNA barcoding of Chlorarachniophytes using nucleomorph ITS sequences. J. Phycol. 46: 743-750.
Guiry, M.D. (1992). Species concepts in marine red algae. Prog. Phycol. Res. 8: 251-278.
Guiry, M.D. & Guiry, G.M. (2014). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org (searched on 24 January 2014).
Gutner-Hoch, E. & Fine, M. (2011). Genotypic diversity and distribution of Ostreobium quekettii within scleractinian corals. Coral Reefs 30: 643-650.
Hagino, K., Bendif, E.M., Young, J.R., Kogame, K., Probert, I., Takano, Y., Horiguchi, T., de Vargas, C., & Okada, H. (2011). New evidence for morphological and genetic variation in the cosmopolitan coccolithophore Emiliania huxleyi (Prymnesiophyceae) from the cox1b-atp4 genes. J. Phycol. 47: 1164-1176.
Hall, J.D., Fucikova, K., Lo, C., Lewis, L.A., & Karol, K.G. (2010). An assessment of proposed DNA barcodes in freshwater green algae. Cryptogam. Algol. 31: 529-555.
Hamsher, S.E., Evans, K.M., Mann, D.G., Poulickova, A., & Saunders, G.W. (2011). Barcoding diatoms: Exploring alternatives to COI-5P. Protist 162: 405-422.
Hare, M.P. (2001). Prospects for nuclear gene phylogeography. Trends Ecol. Evol. 16: 700-706.
Harrington, R.C. & Near, T.J. (2012). Phylogenetic and coalescent strategies of species delimitation in Snubnose Darters (Percidae: Etheostoma). Syst. Biol. 61: 63-79.
Hart, M.W. & Sunday, J. (2007). Things fall apart: biological species form unconnected parsimony networks. Biol. Lett. 3: 509-512.
Hausdorf, B. (2011). Progress toward a general species concept. Evolution 65: 923-931.
Hebert, P.D.N., Cywinska, A., Ball, S.L., & deWaard, J.R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. B 270: 313-321.
Hebert, P.D.N., Stoeckle, M.Y., Zemlak, T.S., & Francis, C.M. (2004). Identification of birds through DNA barcodes. PLoS Biol. 2: e312.
Hey, J. (2006). On the failure of modern species concepts. Trends Ecol. Evol. 21: 447-450.
Hey, J. & Pinho, C. (2012). Population genetics and objectivity in species diagnosis. Evolution 66: 1413-1429.
Hickerson, M.J., Meyer, C.P., & Moritz, C. (2006). DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 55: 729-739.
Hind, K.R. & Saunders, G.W. (2013). Molecular markers from three organellar genomes unravel complex taxonomic relationships within the coralline algal genus Chiharaea (Corallinales, Rhodophyta). Mol. Phylogenet. Evol. 67: 529-540.
Hiraoka, M., Ichihara, K., Zhu, W., Ma, J., & Shimada, S. (2011). Culture and hybridization experiments on an Ulva clade including the Qingdao strain blooming in the Yellow Sea. PLoS One 6: e19371.
Hoef-Emden, K. (2007). Revision of the genus Cryptomonas (Cryptophyceae) II: incongruences between the classical morphospecies concept and molecular phylogeny in smaller pyrenoid-less cells. Phycologia 46: 402-428.
Hoef-Emden, K. (2012). Pitfalls of establishing DNA barcoding systems in protists: The Cryptophyceae as a test case. PLoS One 7: e43652.
Hudson, R.R. & Coyne, J.A. (2002). Mathematical consequences of the genealogical species concept. Evolution 56: 1557-1565.
Janouškovec, J., Liu, S.-L., Martone, P.T., Carré, W., Leblanc, C., Collén, J., & Keeling, P.J. (2013). Evolution of red algal plastid genomes: Ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One 8: e59001.
John, D.M. & Maggs, C.A. (1997). Species problems in eukaryotic algae: a modern perspective. In Species: The Units of Biodiversity (Claridge, M.F., Dawah, H.A., & Wilson, M.R., editors), 83-107. Chapman & Hall, London.
Kamiya, M. (2004). Speciation and biogeography of the Caloglossa leprieurii complex (Delesseriaceae, Rhodophyta). J. Plant Res. 117: 421-428.
Kermarrec, L., Franc, A., Rimet, F., Chaumeil, P., Humbert, J.F., & Bouchez, A. (2013). Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: a test for freshwater diatoms. Mol. Ecol. Resour. 13: 607-619.
Kim, J.I., Shin, W., & Triemer, R.E. (2013). Cryptic speciation in the genus Cryptoglena (Euglenaceae) revealed by nuclear and plastid SSU and LSU rRNA gene. J. Phycol. 49: 92-102.
Kizirian, D. & Donnelly, M. (2004). The criterion of reciprocal monophyly and classification of nested diversity at the species level. Mol. Phylogenet. Evol. 32: 1072-1076.
Klein, J., Sato, A., Nagl, S., & O’hUigín, C. (1998). Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst. 29: 1-21.
Klopper, S., John, U., Zingone, A., Mangoni, O., Kooistra, W., & Cembella, A.D. (2013). Phylogeny and morphology of a Chattonella (Raphidophyceae) species from the Mediterranean Sea: what is C. subsalsa? Eur. J. Phycol. 48: 79-92.
Knowles, L.L. & Carstens, B.C. (2007). Delimiting species without monophyletic gene trees. Syst. Biol. 56: 887-895.
Krienitz, L. & Bock, C. (2012). Present state of the systematics of planktonic coccoid green algae of inland waters. Hydrobiologia 698: 295-326.
Kucera, H. & Saunders, G.W. (2008). Assigning morphological variants of Fucus (Fucales, Phaeophyceae) in Canadian waters to recognized species using DNA barcoding. Botany 86: 1065-1079.
Kucera, H. & Saunders, G.W. (2012). A survey of Bangiales (Rhodophyta) based on multiple molecular markers reveals cryptic diversity. J. Phycol. 48: 869-882.
Kusber, W.-H. & Jahn, R. (2003). Annotated list of diatom names by Horst Lange-Bertalot and co-workers. Version 3.0. http://www.algaterra.org/names_version3_0.pdf (accessed 13 Feb 2014).
Kynčlová, A., Škaloud, P., & Škaloudová, M. (2010). Unveiling hidden diversity in the Synura petersenii species complex (Synurophyceae, Heterokontophyta). Nova Hedwigia Beih. 136: 283-298.
LaJeunesse, T.C., Parkinson, J.E., & Reimer, J.D. (2012). A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J. Phycol. 48: 1380-1391.
LaJeunesse, T.C. & Thornhill, D.J. (2011). Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS One 6: e29013.
Lam, D.W., Entwisle, T.J., Eloranta, P., Kwandrans, J., & Vis, M.L. (2012). Circumscription of species in the genus Sirodotia (Batrachospermales, Rhodophyta) based on molecular and morphological data. Eur. J. Phycol. 47: 42-50.
Lane, C.E., Lindstrom, S.C., & Saunders, G.W. (2007). A molecular assessment of northeast Pacific Alaria species (Laminariales, Phaeophyceae) with reference to the utility of DNA barcoding. Mol. Phylogenet. Evol. 44: 634-648.
Lange, M., Chen, Y.Q., & Medlin, L.K. (2002). Molecular genetic delineation of Phaeocystis species (Prymnesiophyceae) using coding and non-coding regions of nuclear and plastid genomes. Eur. J. Phycol. 37: 77-92.
Le Gall, L. & Saunders, G.W. (2010). DNA barcoding is a powerful tool to uncover algal diversity: a case study of the Phyllophoraceae (Gigartinales, Rhodophyta) in the Canadian flora. J. Phycol. 46: 374-389.
Leaché, A.D. & Fujita, M.K. (2010). Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proc. R. Soc. B 277: 3071-3077.
Leaché, A.D., Harris, R.B., Rannala, B., & Yang, Z. (2014). The influence of gene flow on species tree estimation: A simulation study. Syst. Biol. 63: 17-30.
Leavitt, S.D., Fankhauser, J.D., Leavitt, D.H., Porter, L.D., Johnson, L.A., & St. Clair, L.L. (2011). Complex patterns of speciation in cosmopolitan ‘‘rock posy’’ lichens – Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol. Phylogenet. Evol. 59: 587-602.
Leliaert, F., Verbruggen, H., Wysor, B., & De Clerck, O. (2009). DNA taxonomy in morphologically plastic taxa: Algorithmic species delimitation in the Boodlea complex (Chlorophyta: Cladophorales). Mol. Phylogenet. Evol. 53: 122-133.
Lelong, A., Hegaret, H., Soudant, P., & Bates, S.S. (2012). Pseudo-nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: revisiting previous paradigms. Phycologia 51: 168-216.
Lemmon, A.R., Emme, S.A., & Lemmon, E.M. (2012). Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61: 727-744.
Lin, S., Zhang, H., Hou, Y., Zhuang, Y., & Miranda, L. (2009). High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob genes for dinoflagellate DNA barcoding. Appl. Environ. Microbiol. 75: 1279-1290.
Litaker, R.W., Vandersea, M.W., Faust, M.A., Kibler, S.R., Chinain, M., Holmes, M.J., Holland, W.C., & Tester, P.A. (2009). Taxonomy of Gambierdiscus including four new species, Gambierdiscus caribaeus, Gambierdiscus carolinianus, Gambierdiscus carpenteri and Gambierdiscus ruetzleri (Gonyaulacales, Dinophyceae). Phycologia 48: 344-390.
Litaker, R.W., Vandersea, M.W., Kibler, S.R., Reece, K.S., Stokes, N.A., Lutzoni, F.M., Yonish, B.A., West, M.A., Black, M.N.D., & Tester, P.A. (2007). Recognizing dinoflagellate species using ITS rDNA sequences. J. Phycol. 43: 344-355.
Liu, H., Probert, I., Uitz, J., Claustre, H., Aris-Brosou, S., Frada, M., Not, F., & de Vargas, C. (2009). Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc. Natl Acad. Sci. U.S.A. 106: 12803-12808.
Logares, R., Rengefors, K., Kremp, A., Shalchian-Tabrizi, K., Boltovskoy, A., Tengs, T., Shurtleff, A., & Klaveness, D. (2007). Phenotypically different microalgal morphospecies with identical ribosomal DNA: A case of rapid adaptive evolution? Microb. Ecol. 53: 549-561.
Lundholm, N., Bates, S.S., Baugh, K.A., Bill, B.D., Connell, L.B., Léger, C., & Trainer, V.L. (2012). Cryptic and pseudo-cryptic diversity in diatoms – with descriptions of Pseudo-nitzschia hasleana sp. nov. and P. fryxelliana sp. nov. J. Phycol. 48: 436-454.
Lundholm, N., Moestrup, O., Kotaki, Y., Hoef-Emden, K., Scholin, C., & Miller, P. (2006). Inter- and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J. Phycol. 42: 464-481.
Luo, W., Proschold, T., Bock, C., & Krienitz, L. (2010). Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biol. 12: 545-553.
Lurling, M. (2003). Phenotypic plasticity in the green algae Desmodesmus and Scenedesmus with special reference to the induction of defensive morphology. Ann. Limnol. 39: 85-101.
Macaya, E.C. & Zuccarello, G.C. (2010). DNA barcoding and genetic divergence in the giant kelp macrocystis (Laminariales). J. Phycol. 46: 736-742.
Maddison, W.P. (1997). Gene trees in species trees. Syst. Biol. 46: 523-536.
Maddison, W.P. & Knowles, L.L. (2006). Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55: 21-30.
Maggs, C.A., Fletcher, H.L., Fewer, D., Loade, L., Mineur, F., & Johnson, M.P. (2011). Speciation in red algae: Members of the Ceramiales as model organisms. Integr. Comp. Biol. 51: 492-504.
Mailund, T. (2009). On gene trees and species trees. Available at http://www.mailund.dk/index.php/2009/02/12/on-gene-trees-and-species-trees/ (last accessed 22 Jan 2014).
Mallet, J. (1995). A species definition for the modern synthesis. Trends Ecol. Evol. 10: 294-299.
Manhart, J.R. & McCourt, R.M. (1992). Molecular data and species concepts in the algae. J. Phycol. 28: 730-737.
Mann, D.G. (1999). The species concept in diatoms. Phycologia 38: 437-495.
Mann, D.G. (2010). Discovering diatom species: is a long history of disagreements about species-level taxonomy now at an end? Plant. Ecol. Evol. 143: 251-264.
Mann, D.G., Sato, S., Trobajo, R., Vanormelingen, P., & Souffreau, C. (2010). DNA barcoding for species identification and discovery in diatoms. Cryptogam. Algol. 31: 557-577.
Masters, B.C., Fan, V., & Ross, H.A. (2011). Species delimitation – a geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 11: 154-157.
Mattio, L. & Payri, C. (2010). Assessment of five markers as potential barcodes for identifying Sargassum subgenus Sargassum species (Phaeophyceae, Fucales). Cryptogam. Algol. 31: 467-485.
Mayden, R.L. (1997). A hierarchy of species concepts: the denouement in the saga of the species problem. In Species: The Units of Biodiversity (Claridge, M.F., Dawah, H.A., & Wilson, M.R., editors), 381-424. Chapman and Hall, London.
Mayr, E. (1996). What is a species, and what is not? Philos. Sci. 63: 262-277.
Mayr, E. & Ashlock, P.D. (1991). Principles of systematic zoology. McGraw-Hill, New York.
McCormack, J.E., Hird, S.M., Zellmer, A.J., Carstens, B.C., & Brumfield, R.T. (2013). Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol. 66: 526-538.
McCormack, J.E., Maley, J.M., Hird, S.M., Derryberry, E.P., Graves, G.R., & Brumfield, R.T. (2012). Next-generation sequencing reveals phylogeographic structure and a species tree for recent bird divergences. Mol. Phylogenet. Evol. 62: 397-406.
McDevit, D.C. & Saunders, G.W. (2009). On the utility of DNA barcoding for species differentiation among brown macroalgae (Phaeophyceae) including a novel extraction protocol. Phycol. Res. 57: 131-141.
McManus, H.A. & Lewis, L.A. (2011). Molecular phylogenetic relationships in the freshwater family Hydrodictyaceae (Sphaeropleales, Chlorophyceae), with an emphasis on Pediastrum duplex. J. Phycol. 47: 152-163.
Meier, R., Shiyang, K., Vaidya, G., & Ng, P.K.L. (2006). DNA barcoding and taxonomy in diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 55: 715-728.
Meyer, C.P. & Paulay, G. (2005). DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 3: 2229-2238.
Milstein, D. & Saunders, G.W. (2012). DNA barcoding of Canadian Ahnfeltiales (Rhodophyta) reveals a new species – Ahnfeltia borealis sp. nov. Phycologia 51: 247-259.
Mols-Mortensen, A., Neefus, C.D., Nielsen, R., Gunnarsson, K., Egilsdóttir, S., Pedersen, P.M., & Brodie, J. (2012). New insights into the biodiversity and generic relationships of foliose Bangiales (Rhodophyta) in Iceland and the Faroe Islands. Eur. J. Phycol. 47: 146-159.
Monaghan, M.T., Wild, R., Elliot, M., Fujisawa, T., Balke, M., Inward, D.J., Lees, D.C., Ranaivosolo, R., Eggleton, P., Barraclough, T.G., & Vogler, A.P. (2009). Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58: 298-311.
Moniz, M.B.J. & Kaczmarska, I. (2010). Barcoding of diatoms: nuclear encoded ITS revisited. Protist 161: 7-34.
Moniz, M.B.J., Rindi, F., Novis, P.M., Broady, P.A., & Guiry, M.D. (2012). Molecular phylogeny of Antarctic Prasiola (Prasiolales, Trebouxiophyceae) reveals extensive cryptic diversity. J. Phycol. 48: 940-955.
Moon-van der Staay, S.Y., De Wachter, R., & Vaulot, D. (2001). Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409: 607-610.
Müller, T., Philippi, N., Dandekar, T., Schultz, J., & Wolf, M. (2007). Distinguishing species. RNA 13: 1469-1472.
Neustupa, J., Šťastný, J., Nemjová, K., Mazalová, P., Goodyer, E., Poulíčková, A., & Škaloud, P. (2011). A novel, combined approach to assessing species delimitation and biogeography within the well-known desmid species Micrasterias fimbriata and M. rotata (Desmidiales, Steptophyta). Hydrobiologia 667: 223-239.
Ni-Ni-Win, Hanyuda, T., Draisma, S.G.A., Furnari, G., Meinesz, A., & Kawai, H. (2011). Padina ditristromatica sp. nov. and Padina pavonicoides sp. nov. (Dictyotales, Phaeophyceae), two new species from the Mediterranean Sea based on morphological and molecular markers. Eur. J. Phycol. 46: 327-341.
Niemiller, M.L., Near, T.J., & Fitzpatrick, B.M. (2012). Delimiting species using multilocus data: Diagnosing cryptic diversity in the Southern Cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae). Evolution 66: 846-866.
Niwa, K., Iida, S., Kato, A., Kawai, H., Kikuchi, N., Kobiyama, A., & Aruga, Y. (2009). Genetic diversity and introgression in two cultivated species (Porphyra yezoensis and Porphyra tenera) and closely related wild species of Porphyra (Bangiales, Rhodophyta). J. Phycol. 45: 493-502.
Nosil, P., Funk, D.J., & Ortiz-Barrientos, D. (2009). Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18: 375-402.
Not, F., Siano, R., Kooistra, W.H.C.F., Simon, N., Vaulot, D., & Probert, I. (2012). Diversity and ecology of eukaryotic marine phytoplankton. Adv. Bot. Res. 64: 1-53.
O’Meara, B.C. (2010). New heuristic methods for joint species delimitation and species tree inference. Syst. Biol. 59: 59-73.
Palumbi, S.R., Cipriano, F., & Hare, M.P. (2001). Predicting nuclear gene coalescence from mitochondrial data: The three-times rule. Evolution 55: 859-868.
Pamilo, P. & Nei, M. (1988). Relationships between gene trees and species trees. Mol. Biol. Evol. 5: 568-583.
Pawlowski, J., Audic, S., Adl, S., Bass, D., Belbahri, L., Berney, C., Bowser, S.S., Cepicka, I., Decelle, J., Dunthorn, M., Fiore-Donno, A.M., Gile, G.H., Holzmann, M., Jahn, R., Jirku, M., Keeling, P.J., Kostka, M., Kudryavtsev, A., Lara, E., Lukes, J., Mann, D.G., Mitchell, E.A.D., Nitsche, F., Romeralo, M., Saunders, G.W., Simpson, A.G.B., Smirnov, A.V., Spouge, J.L., Stern, R.F., Stoeck, T., Zimmermann, J., Schindel, D., & de Vargas, C. (2012). CBOL Protist Working Group: Barcoding Eukaryotic Richness beyond the Animal, Plant, and Fungal Kingdoms. PLoS Biol. 10: e1001419.
Pawlowski, J. & Holzmann, M. (2009). Diversity and geographic distribution of benthic foraminifera: a molecular perspective. In Protist Diversity and Geographical Distribution (8) (Foissner, W. & Hawksworth, D., editors), 83-94. Springer Netherlands.
Payo, D.A., Leliaert, F., Verbruggen, H., D’Hondt, S., Calumpong, H.P., & De Clerck, O. (2013). Extensive cryptic species diversity and fine-scale endemism in the marine red alga Portieria in the Philippines. Proc. R. Soc. B 280: 20122660.
Peters, A.F., Van Wijk, S.J., Cho, G.Y., Scornet, D., Hanyuda, T., Kawai, H., Schroeder, D.C., Cock, J.M., & Boo, S.M. (2010). Reinstatement of Ectocarpus crouaniorum Thuret in Le Jolis as a third common species of Ectocarpus (Ectocarpales, Phaeophyceae) in Western Europe, and its phenology at Roscoff, Brittany. Phycol. Res. 58: 157-170.
Piganeau, G., Eyre-Walker, A., Grimsley, N., & Moreau, H. (2011). How and why DNA barcodes underestimate the diversity of microbial eukaryotes. PLoS One 6: e16342.
Pons, J., Barraclough, T.G., Gomez-Zurita, J., Cardoso, A., Duran, D.P., Hazell, S., Kamoun, S., Sumlin, W.D., & Vogler, A.P. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55: 595-609.
Posada, D. & Crandall, K.A. (2001). Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 16: 37-45.
Powell, J.R. (2012). Accounting for uncertainty in species delineation during the analysis of environmental DNA sequence data. Methods Ecol. Evol. 3: 1-11.
Pröschold, T., Darienko, T., Silva, P.C., Reisser, W., & Krienitz, L. (2011). The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 13: 350-364.
Pröschold, T., Marin, B., Schlosser, U.G., & Melkonian, M. (2001). Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152: 265-300.
Puillandre, N., Lambert, A., Brouillet, S., & Achaz, G. (2011). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21: 1864-1877.
Reid, N. & Carstens, B. (2012). Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol. Biol. 12: 196.
Rindi, F., Mikhailyuk, T.I., Sluiman, H.J., Friedl, T., & López-Bautista, J.M. (2011). Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta). Mol. Phylogenet. Evol. 58: 218-231.
Robba, L., Russell, S.J., Barker, G.L., & Brodie, J. (2006). Assessing the use of the mitochondrial cox1 marker for use in DNA barcoding of red algae (Rhodophyta). Am. J. Bot. 93: 1101-1108.
Roe, A.D., Rice, A.V., Bromilow, S.E., Cooke, J.E.K., & Sperling, F.A.H. (2010). Multilocus species identification and fungal DNA barcoding: insights from blue stain fungal symbionts of the mountain pine beetle. Mol. Ecol. Resour. 10: 946-959.
Rosenberg, N.A. & Nordborg, M. (2002). Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 3: 380-390.
Rybalka, N., Andersen, R.A., Kostikov, I., Mohr, K.I., Massalski, A., Olech, M., & Friedl, T. (2009). Testing for endemism, genotypic diversity and species concepts in Antarctic terrestrial microalgae of the Tribonemataceae (Stramenopiles, Xanthophyceae). Environ. Microbiol. 11: 554-565.
Rybalka, N., Wolf, M., Andersen, R., & Friedl, T. (2013). Congruence of chloroplast- and nuclear-encoded DNA sequence variations used to assess species boundaries in the soil microalga Heterococcus (Stramenopiles, Xanthophyceae). BMC Evol. Biol. 13: 39.
Saez, A.G., Probert, I., Geisen, M., Quinn, P., Young, J.R., & Medlin, L.K. (2003). Pseudo-cryptic speciation in coccolithophores. Proc. Natl Acad. Sci. U.S.A. 100: 7163-7168.
Sampayo, E.M., Dove, S., & Lajeunesse, T.C. (2009). Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus Symbiodinium. Mol. Ecol. 18: 500-519.
Sato, S., Nishimura, T., Uehara, K., Sakanari, H., Tawong, W., Hariganeya, N., Smith, K., Rhodes, L., Yasumoto, T., Taira, Y., Suda, S., Yamaguchi, H., & Adachi, M. (2011). Phylogeography of Ostreopsis along West Pacific coast, with special reference to a novel clade from Japan. PLoS One 6: e27983.
Saunders, G.W. (2005). Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Phil. Trans. R. Soc. B 360: 1879-1888.
Saunders, G.W. & Kucera, H. (2010). An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogam. Algol. 31: 487-528.
Saunders, G.W. & McDevit, D.C. (2012). Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms. Methods Mol. Biol. 858: 207-222.
Schmidt, M., Horn, S., Flieger, K., Ehlers, K., Wilhelm, C., & Schnetter, R. (2012). Synchroma pusillum sp. nov. and other new algal isolates with chloroplast complexes confirm the Synchromophyceae (Ochrophyta) as a widely distributed group of amoeboid algae. Protist 163: 544-559.
Serrao, E.A., Alice, L.A., & Brawley, S.H. (1999). Evolution of the Fucaceae (Phaeophyceae) inferred from nrDNA-ITS. Journal of Phycology 35: 382-394.
Shaffer, H.B. & Thomson, R.C. (2007). Delimiting species in recent radiations. Syst. Biol. 56: 896-906.
Shaw, K.L. (1998). Species and the diversity of natural groups. In Endless Forms: Species and Speciation (Howard, D.J. & Berlocher, S.H., editors), 44-56. Oxford University Press, Oxford.
Sherwood, A.R. & Presting, G.G. (2007). Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J. Phycol. 43: 605-608.
Sherwood, A.R., Sauvage, T., Kurihara, A., Conklin, K.Y., & Presting, G.G. (2010). A comparative analysis of COI, LSU and UPA marker data for the Hawaiian florideophyte Rhodophyta: implications for DNA barcoding of red algae. Cryptogam. Algol. 31: 451-465.
Sherwood, A.R., Vis, M.L., Entwisle, T.J., Necchi Jr, O., & Presting, G.G. (2008). Contrasting intra versus interspecies DNA sequence variation for representatives of the Batrachospermales (Rhodophyta): Insights from a DNA barcoding approach. Phycol. Res. 56: 269-279.
Silberfeld, T., Bittner, L., Fernández-García, C., Cruaud, C., Rousseau, F., de Reviers, B., Leliaert, F., Payri, C.E., & De Clerck, O. (2013). Species diversity, phylogeny and large scale biogeographic patterns of the genus Padina (Phaeophyceae, Dictyotales). J. Phycol. 49: 130-142.
Simpson, G.G. (1951). The species concept. Evol. Dev. 5: 285-298.
Sites, J.W. & Marshall, J.C. (2003). Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 18: 462-470.
Škaloud, P., Kynclova, A., Benada, O., Kofronova, O., & Skaloudova, M. (2012). Toward a revision of the genus Synura, section Petersenianae (Synurophyceae, Heterokontophyta): morphological characterization of six pseudo-cryptic species. Phycologia 51: 303-329.
Škaloud, P. & Peksa, O. (2010). Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta). Mol. Phylogenet. Evol. 54: 36-46.
Škaloud, P. & Rindi, F. (2013). Ecological differentiation of cryptic species within an asexual protist morphospecies: A case study of filamentous green alga Klebsormidium (Streptophyta). J. Eukaryot. Microbiol. 60: 350-362.
Šlapeta, J., Lopez-Garcia, P., & Moreira, D. (2006). Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol. Biol. Evol. 23: 23-29.
Soehner, S., Zinssmeister, C., Kirsch, M., & Gottschling, M. (2012). Who am I – and if so, how many? Species diversity of calcareous dinophytes (Thoracosphaeraceae, Peridiniales) in the Mediterranean Sea. Org. Divers. Evol. 12: 339-348.
Souffreau, C., Vanormelingen, P., Van de Vijver, B., Isheva, T., Verleyen, E., Sabbe, K., & Vyverman, W. (2013). Molecular evidence for distinct Antarctic lineages in the cosmopolitan terrestrial diatoms Pinnularia borealis and Hantzschia amphioxys. Protist 164: 101-115.
Stern, R.F., Andersen, R.A., Jameson, I., Küpper, F.C., Coffroth, M.-A., Vaulot, D., Le Gall, F., Véron, B., Brand, J.J., Skelton, H., Kasai, F., Lilly, E.L., & Keeling, P.J. (2012). Evaluating the ribosomal Internal Transcribed Spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One 7: e42780.
Stern, R.F., Horak, A., Andrew, R.L., Coffroth, M.-A., Andersen, R.A., Küpper, F.C., Jameson, I., Hoppenrath, M., Véron, B., Kasai, F., Brand, J., James, E.R., & Keeling, P.J. (2010). Environmental barcoding reveals massive dinoflagellate diversity in marine environments. PLoS One 5: e13991.
Subirana, L., Péquin, B., Michely, S., Escande, M.-L., Meilland, J., Derelle, E., Marin, B., Piganeau, G., Desdevises, Y., Moreau, H., & Grimsley, N.H. (2013). Morphology, genome plasticity, and phylogeny in the genus Ostreococcus reveal a cryptic species, O. mediterraneus sp. nov. (Mamiellales, Mamiellophyceae). Protist 164: 643-659.
Sutherland, J.E., Lindstrom, S.C., Nelson, W.A., Brodie, J., Lynch, M.D.J., Hwang, M.S., Choi, H.-G., Miyata, M., Kikuchi, N., Oliveira, M.C., Farr, T., Neefus, C., Mols-Mortensen, A., Milstein, D., & Müller, K.M. (2011). A new look at an ancient order: generic revision of the Bangiales (Rhodophyta). J. Phycol. 47: 1131-1151.
Sym, S.D., Pienaar, R.N., Edvardsen, B., & Egge, E.S. (2011). Fine structure and systematics of Prymnesium radiatum sp. nov. (Prymnesiophyceae) from False Bay and Franskraal, South Africa. Eur. J. Phycol. 46: 229-248.
Tautz, D., Arctander, P., Minelli, A., Thomas, R.H., & Vogler, A.P. (2003). A plea for DNA taxonomy. Trends Ecol. Evol. 18: 70-74.
Taylor, J.W., Jacobson, D.J., Kroken, S., Kasuga, T., Geiser, D.M., Hibbett, D.S., & Fisher, M.C. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31: 21-32.
Templeton, A.R. (2001). Using phylogeographic analyses of gene trees to test species status and processes. Mol. Ecol. 10: 779-791.
Trainor, F.R. (1998). Biological aspects of Scenedesmus (Chlorophyceae) phenotypic plasticity. Nova Hedwigia Beih. 117: 1-367.
Tronholm, A., Leliaert, F., Sanson, M., Afonso-Carrillo, J., Tyberghein, L., Verbruggen, H., & De Clerck, O. (2012). Contrasting geographical distributions as a result of thermal tolerance and long-distance dispersal in two allegedly widespread tropical brown algae. PLoS One 7: e30813.
Tronholm, A., Steen, F., Tyberghein, L., Leliaert, F., Verbruggen, H., Siguan, M.A.R., & De Clerck, O. (2010). Species delimitation, taxonomy, and biogeography of Dictyota in Europe (Dictyotales, Phaeophyceae). J. Phycol. 46: 1301-1321.
van der Strate, H.J., Boele-Bos, S.A., Olsen, J.L., van de Zande, L., & Stam, W.T. (2002). Phylogeographic studies in the tropical seaweed Cladophoropsis membranacea (Chlorophyta, Ulvophyceae) reveal a cryptic species complex. J. Phycol. 38: 572-582.
Vanelslander, B., Créach, V., Vanormelingen, P., Ernst, A., Chepurnov, V.A., Sahan, E., Muyzer, G., Stal, L.J., Vyverman, W., & Sabbe, K. (2009). Ecological differentiation between sympatric pseudocryptic species in the estuarine benthic diatom Navicula phyllepta (Bacillariophyceae). J. Phycol. 45: 1278-1289.
Vanormelingen, P., Chepurnov, V.A., Mann, D.G., Sabbe, K., & Vyverman, W. (2008). Genetic divergence and reproductive barriers among morphologically heterogeneous sympatric clones of Eunotia bilunaris sensu lato (Bacillariophyta). Protist 159: 73-90.
Vanormelingen, P., Hegewald, E., Braband, A., Kitschke, M., Friedl, T., Sabbe, K., & Vyverman, W. (2007). The systematics of a small spineless Desmodesmus species, D. constato-granulatus (Sphaeropleales, Chlorophyceae), based on ITS2 rDNA sequence analyses and cell wall morphology. J. Phycol. 43: 378-396.
Verbruggen, H. (2014). Morphological complexity, plasticity, and species diagnosability in the application of old species names in DNA-based taxonomies. J. Phycol.: in press.
Verbruggen, H., Leliaert, F., Maggs, C.A., Shimada, S., Schils, T., Provan, J., Booth, D., Murphy, S., De Clerck, O., Littler, D.S., Littler, M.M., & Coppejans, E. (2007). Species boundaries and phylogenetic relationships within the green algal genus Codium (Bryopsidales) based on plastid DNA sequences. Mol. Phylogenet. Evol. 44: 240-254.
Verbruggen, H., Vlaeminck, C., Sauvage, T., Sherwood, A.R., Leliaert, F., & De Clerck, O. (2009). Phylogenetic analysis of Pseudochlorodesmis strains reveals cryptic diversity above the family level in the siphonous green algae (Bryopsidales, Chlorophyta). J. Phycol. 45: 726-731.
Wagner, C.E., Keller, I., Wittwer, S., Selz, O.M., Mwaiko, S., Greuter, L., Sivasundar, A., & Seehausen, O. (2013). Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol. Ecol. 22: 787-798.
Walker, R.H., Brodie, J., Russell, S., Irvine, L.M., & Orfanidis, S. (2009). Biodiversity of coralline algae in the northeastern Atlantic including Corallina caespitosa sp. nov. (Corallinoideae, Rhodophyta). J. Phycol. 45: 287-297.
Wiens, J.J. (2007). Species delimitation: New approaches for discovering diversity. Syst. Biol. 56: 875-878.
Wiens, J.J. & Penkrot, T.A. (2002). Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus). Syst. Biol. 51: 69-91.
Wiley, E.O. (1978). The Evolutionary Species Concept reconsidered. Syst. Zool. 21: 17-26.
Wilkins, J.S. (2009). Species: A History of the Idea. University of California Press, Berkeley.
Wolf, M., Chen, S., Song, J., Ankenbrand, M., & Müller, T. (2013). Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences – A proof of concept. PLoS One 8: e66726.
Yang, Z. & Rannala, B. (2010). Bayesian species delimitation using multilocus sequence data. Proc. Natl Acad. Sci. U.S.A. 107: 9264-9269.
Zachos, F.E. & Lovari, S. (2013). Taxonomic inflation and the poverty of the Phylogenetic Species Concept – a reply to Gippoliti and Groves.
Zardi, G.I., Nicastro, K.R., Canovas, F., Ferreira Costa, J., Serrão, E.A., & Pearson, G.A. (2011). Adaptive traits are maintained on steep selective gradients despite gene flow and hybridization in the intertidal zone. PLoS One 6: e19402.
Zimmermann, J., Jahn, R., & Gemeinholzer, B. (2011). Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org. Divers. Evol. 11: 173-192.
Zink, R.M. & Barrowclough, G.F. (2008). Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 17: 2107-2121.
Zuccarello, G.C. & Lokhorst, G.M. (2005). Molecular phylogeny of the genus Tribonema (Xanthophyceae) using rbcL gene sequence data: monophyly of morphologically simple algal species. Phycologia 44: 384-392.
Zuccarello, G.C., Schidlo, N., McIvor, L., & Guiry, M.D. (2005). A molecular re-examination of speciation in the intertidal red alga Mastocarpus stellatus (Gigartinales, Rhodophyta) in Europe. Eur. J. Phycol. 40: 337-344.
Zuccarello, G.C. & West, J.A. (2003). Multiple cryptic species: Molecular diversity and reproductive isolation in the Bostrychia radicans/B. moritziana complex (Rhodomelaceae, Rhodophyta) with focus on North American isolates. J. Phycol. 39: 948-959.
179-196

One thought on “DNA-based species delimitation in algae”